
# are repository servers and will give you the ability to manage multiple # Chocolatey Software recommends Nexus, Artifactory Pro, or ProGet as they # generally really quick to set up and there are quite a few options. # You'll need an internal/private cloud repository you can use. Internal/Private Cloud Repository Set Up # # Here are the requirements necessary to ensure this is successful. Your use of the packages on this site means you understand they are not supported or guaranteed in any way. With any edition of Chocolatey (including the free open source edition), you can host your own packages and cache or internalize existing community packages. Packages offered here are subject to distribution rights, which means they may need to reach out further to the internet to the official locations to download files at runtime.įortunately, distribution rights do not apply for internal use.
If you are an organization using Chocolatey, we want your experience to be fully reliable.ĭue to the nature of this publicly offered repository, reliability cannot be guaranteed.
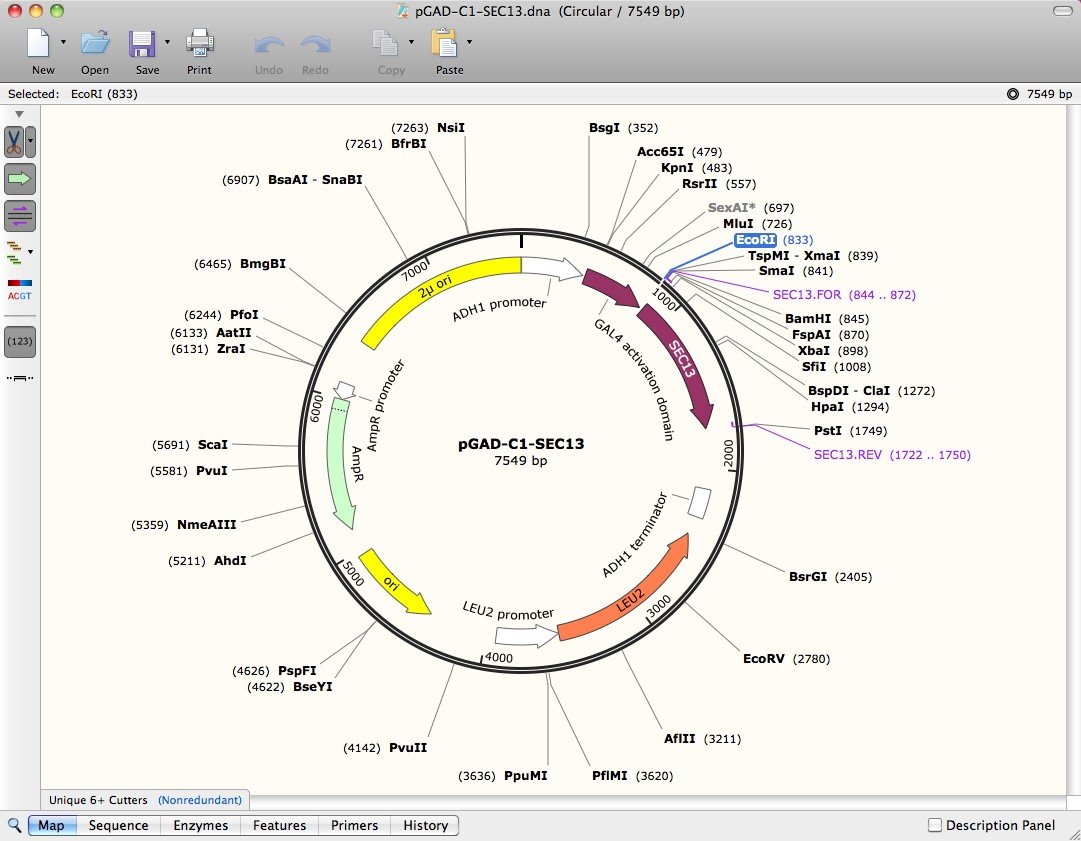

However, my PI said something about being concerned that these enzymes didn't cut 100% so she wants to try a different one that will just linearize by cutting inside the promoter? I don't quite understand, but this tool will be useful to me regardless. One of the promoters would be about 400bp in size, while the other would be about 1400. I can cut with enzymes just outside of each of the promoters as we designed them that way. So ideally I'd like a list of enzymes which are unique to one plasmid or the other. The restriction enzyme digest is primarily to confirm the purity of the plasmid isolated from specific colonies. I am dealing with two highly similar plasmids, of similar size (7300 vs 6400) which differ in the promoter used. I am attempting to find a list of enzymes that cut one plasmid, but not another.


 0 kommentar(er)
0 kommentar(er)
